Crop wild relatives and genetic resources for food security
Population growth and global change are the two major challenges for food security. The human population has increased from one to seven billion during the past 200 years and is expected to reach 11 billion by the end of this century. However, agricultural production is decreasing by 2% each decade due to global change. To meet the Sustainable Development Goal targets by 2030 to bring about zero hunger, we need to boost our food grain production by about 70% and for this we need crops with higher yields, nutritional values, and ability to resist diseases and adapt to changing environments.
Domesticated crops have been under human selection pressure for centuries and their gene pool is limited by the domestication bottleneck. The main aim of the use case is to develop a prototype digital twin that enables the search for CWR and traditional cultivars containing novel genetic resources for improvements of domesticated crops. To start with this, current focus is given to improve the nutritional quality of a climate smart crop, grasspea (Lathyrus sativus L.).
Grasspea is a nutrition rich legume known for resisting drought, extreme temperature and poor soil. Through symbiotic association with Rhizobium bacteria, it fixes nitrogen and thus can grow even on degraded lands. It uses residual moisture for growth and is usually cropped after the main cropping seasons. If other crops fail, the same farms can be covered by it and it is usually considered a lifesaver in developing tropical and subtropical countries. Nevertheless, it presents a fascinating paradox, i.e. it contains a neurotoxin that causes permanent paralysis of lower limbs in adults and brain damage in children, if consumed over longer period as a major diet. It is quite widely distributed and it has about 180 wild relatives meaning huge genetic resources for its improvement.
The toxicity of grasspea is affected by different factors such water stress, soil zinc content, salinity and day length or number of light hours per day during the growing season. Toxicity increases with water and zinc deficiency and with number of day light hours and salinity. This means the same genotype may present different levels of toxicity under different environmental conditions, complicating the matter. We hypothesize that
- grasspea landraces and wild relatives growing in dry areas and in zinc deficient and acidic soils are most likely efficient in water, zinc and sodium uptakes, respectively; and
- through improving these efficacies of grasspea, it is possibly to minimize the production of the neurotoxin chemical.
Thus, the main aim of this work is modelling the habitat suitability of grasspea wild relatives and identifying genotypes with adaptation to these environmental factors to present potential alleles to improve grasspea.

Crop wild relatives and genetic resources for food security and Digital Twin Models

Thus we aim to model the habitat suitability of grasspea wild relatives and identify genotypes with adaptation to these environmental factors to present potential alleles to improve grasspea as a crop. Habitat suitability of grasspea landraces and CWR will be modelled using MoDGPI (modelling the distributions of germplasms of interest) and provide a map of geographic areas that harbor germplasms of interest. MoDGPI uses different high performing species distribution modelling algorithms such as generalized additive modelling (GAM7), random forest (RF), generalised boosted regression modelling (gbm8), and maximum entropy modelling (MaxEnt9) to produce habitat suitability maps of model targets. The algorithms in MoDGPI function by relating occurrence points to environmental variables to produce habitat suitability maps. Results from all algorithms will be evaluated against test data using area under the ROC curve (AUC) and True Skill Statistics (TSS). Maps from less performing models i.e. with AUC < 0.7 and/or TSS < 0.4 will be dropped and only maps from high performing algorithms and models settings will be kept.
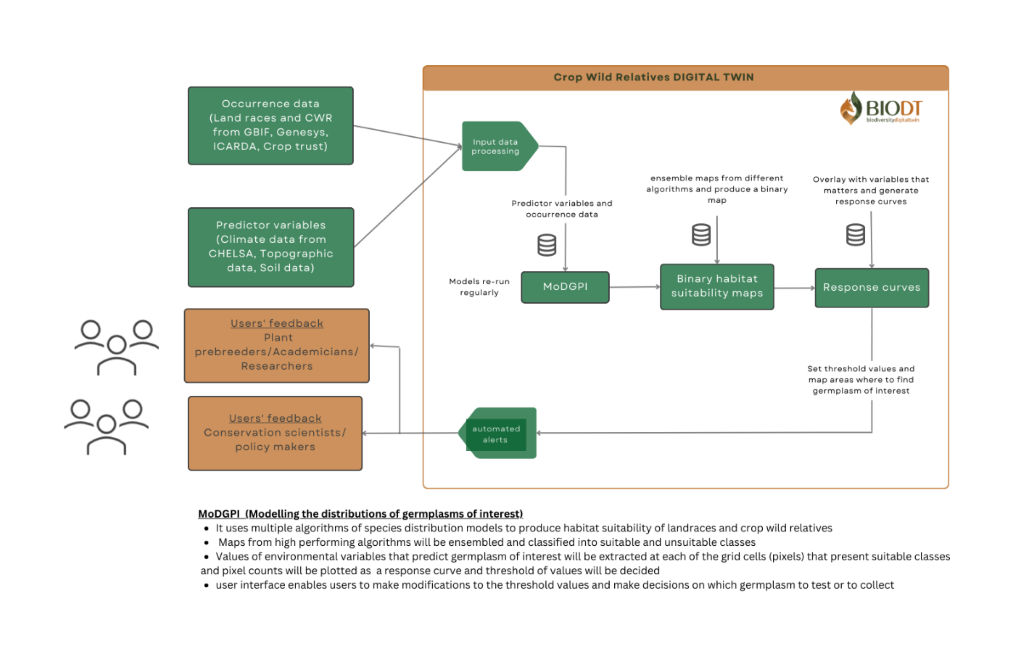
Maps from high performing models will be combined through an ensemble approach and binary maps (maps with suitable and unsuitable classes) will be produced using the maximum sum sensitivity and specificity threshold. The suitable class of the binary maps will be overlaid with a soil zinc map, soil acidity map and rainfall data, and their range of tolerance to these environmental variables will be generated as response curves. Each of the grasspea crop wild relatives will be ranked based on their range of tolerances to these environmental factors. For model targets with high tolerance to these factors, geographic areas where plants presenting the desired genotypes are growing will be mapped and provided. GUI/R shiny interface will be provided to allow user/plant breeders to prioritise the plant genetic resources. The models are also aimed to be built in a way it can be transferable across traits and crops.



